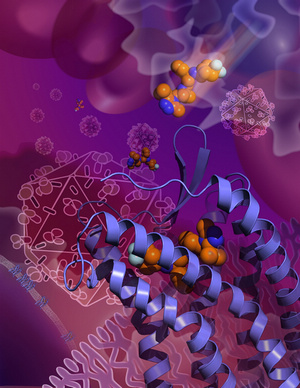
SHANGHAI, CHINA, AND LA JOLLA, CA – September 12, 2013 – In a long-awaited finding, a team of Chinese and US scientists has determined the high-resolution atomic structure of a cell-surface receptor that most strains of HIV use to get into human immune cells. The researchers also showed where maraviroc, an HIV drug, attaches to cells and blocks HIV’s entry. This important breakthough news was released yesterday from Science-AAAS press conference held in Shanghai Institute of Materia Medica, Chinese Academy of Sciences. It is the first time Science-AAAS holding press conference in Shanghai.
“These structural details should help us understand more precisely how HIV infects cells, and how we can do better at blocking that process with next-generation drugs,” said Beili Wu, PhD, professor at the Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences. Wu was the senior investigator for the study, which was published in Science on September 12, 2013.
The study, which focused on the CCR5 receptor, was supported by both US and Chinese research funding agencies. “International collaborations like this one are increasingly needed to solve big problems in science,” said study co-author Raymond C. Stevens, PhD, a professor at The Scripps Research Institute (TSRI) in California. “Now that we have both human CXCR4 and CCR5 HIV co-receptor three-dimensional structures, it is likely we will see the next generation of HIV therapeutics.”
A Major Target
The CCR5 receptor is one of the most sought-after targets for new anti-HIV drugs. Although the AIDS-causing virus was initially discovered to infect cells via another receptor, CD4, researchers found in 1996 that HIV infection also requires a co-receptor—usually CCR5, which sits alongside CD4 on a variety of immune cells.
CCR5’s importance to HIV infection is underscored by the fact that certain genetic variants of it can dramatically raise or lower HIV infection risk, as well as the speed of the disease process after infection. One shortened CCR5 variant, found in about 10 percent of Europeans, is not expressed at all on immune cell surfaces—and people who produce only this variant are almost invulnerable to HIV infection.
Scientists therefore have sought to develop anti-HIV drugs that block the virus from binding to CCR5 or otherwise render the receptor inactive. Yet only a handful of CCR5-inhibiting compounds have been developed so far—and no one knows exactly how they work. “One thing that we’ve lacked is a high-resolution molecular ‘picture’ of the CCR5 receptor structure that we can use for precise drug design,” Wu said.
A Six-Year Quest
Wu came to the Stevens laboratory at TSRI in 2007 to conduct postdoctoral research on the two HIV co-receptors CCR5 and the alternate HIV co-receptor CXCR4. A minority of HIV strains use CXCR4 instead of CCR5 as a co-receptor with CD4 for their initial infiltration of cells.
“Her goal from the beginning was to determine the structures and understand the functions of these two HIV co-receptors and she was very determined which was inspiring,” said Stevens.
Wu spent her postdoctoral years focusing on CXCR4, which posed fewer technical challenges than CCR5. Her landmark study of the CXCR4 structure was published in Science in 2010. After moving back to China to start her own laboratory at SIMM, she returned to the “unfinished business” of the CCR5 structure.
Both CCR5 and CXCR4 belong to a large family of cell receptors known as G protein-coupled receptors (GPCRs). GPCRs are notoriously hard to produce in useful amounts for structural analysis. With their floppy structures, they are also very hard to coax into the ordered, solid lineups of individual molecules—“crystals”—needed for structure determination by X-ray crystallography. Eventually, however, with help from insights gained during the CXCR4 project, Wu as a new professor and her young team of students used a novel “fusion partner” molecule that would hold CCR5 proteins together enough to form usable crystals, together with efforts of computational modeling, compound synthesis and cell signaling assays from Drs. Hualiang Jiang, Hong Liu and Xin Xie’s groups, which led to the structure determination of CCR5.
As in most receptor-structure projects, Wu and her colleagues further stabilized CCR5 with a compound that is known to bind to it, in this case the Pfizer drug maraviroc (sold under the brand name Selzentry®, or Celsentri® outside the US). Marketed for HIV infection since 2007, maraviroc grabs hold of CCR5 in a way that prevents HIV from using the receptor to get into cells. “Maraviroc was thought to lock CCR5 into an inactive conformation, and so we wanted to ‘see’ that conformation at high resolution,” said Wu.
The resulting crystallography data provided that fine-grained picture of CCR5’s HIV-resistant conformation. The data also revealed maraviroc’s precise binding site on CCR5—a site from which the drug molecule clearly influences how the receptor works, even though it is separate from the sites on the receptor that are thought to be used by HIV. The maraviroc binding site is also different from the site used by CCR5’s natural binding-partners, a set of immune proteins called chemokines. Maraviroc thus appears to work against HIV indirectly: not by physically blocking the virus, but by locking the receptor structure into an HIV-insensitive conformation.
“Structural details can offer tremendous insight into how proteins and drugs work, also aiding the development of therapeutic agents,” said Peter Preusch, PhD, of the National Institute of General Medical Sciences, which helped fund the research along with another component of the National Institutes of Health, the National Institute of Allergy and Infectious Diseases. “This study provides knowledge about the interactions between maraviroc and CCR5, a target for anti-HIV therapy, that helps us understand how the drug works at the molecular level and could enable further explorations of HIV biology and approaches to improve drugs targeting such interactions.”
Useful Insights
Comparison of the CCR5 structure with the previously determined CXCR4 structure also provided hints about an important aspect of HIV evolution during infections. Most HIV infections start by using only CCR5 as a co-receptor for cell entry, but in time the virus often switches its co-receptor usage from CCR5 to CXCR4. That opens up more cell types to HIV infection, and the further spread of the virus inside the body is liable to speed up the disease progression towards full-blown AIDS and death.
The new data suggest that the distinction between CCR5 and CXCR4 as co-receptors for HIV infection boils down to relatively subtle differences in structural shapes and electric charge distributions in the HIV binding region—differences that will be of interest to HIV drug developers.
“Knowing the CXCR4 structure and now the CCR5 structure at this level of detail should accelerate the development of drugs that can block HIV by using both of these co-receptors,” said Wu.
She and her colleagues now plan to follow up with structural studies of CCR5 and CXCR4 in complex with the HIV envelope protein gp120 and CD4 to obtain even more informative pictures of the process of viral infection.
Soon after the structure determination of CCR5, SIMM performed structure-based drug design and has obtained several drug lead compounds with more potent antiviral efficacies than maraviroc, which further proves the importance of CCR5 structure on the development of HIV theraputics.
In addition to Wu and Stevens, the contributors to the study, “Structure of the CCR5 Chemokine Receptor – HIV Entry Inhibitor Maraviroc Complex,” were lead author Qiuxiang Tan and Ya Zhu, Jian Li, Zhuxi Chen, Tingting Li, Limin Ma, Jing Li, Wenru Zhang, Xin Xie, Huaiyu Yang, Hualiang Jiang, Hong Liu and Qiang Zhao, all of the Shanghai Institute of Materia Medica; Gye Won Han, Gustavo Fenalti and Vadim Cherezov of TSRI; and Irina Kufareva of the University of California at San Diego.
The study was funded in part by the National Basic Research Program of China (grants 2012CB518000 and 2012CB910400), the National Institutes of Health (R01 AI100604, U54 GM094618 and a supplement to U54 GM094618 as part of the U.S.-China Biomedical Collaborative Research Program), the National Science Foundation of China (grants 31270766, 81161120425, and 81025017) and the Shanghai Science and Technology Committee (grants 11JC1414800 and 12PJ1410500).
Full text: http://www.sciencemag.org/content/early/2013/09/11/science.1241475
Editor comment: http://www.sciencemag.org/content/early/2013/09/11/science.1245384.full