With the big promise of treating eye diseases with cell therapy, retinal stem cells (RSCs), as the cell source, have been the long-lasting research hotspot. In this theme, a central issue is: do human eye actually have retinal stem cells? Interestingly, in lower vertebrates, such as Xenopus and fish, retinal stem cells, residing in ciliary marginal zone (CMZ), actively contribute to the sustainable growth of retinas throughout the organism’s lifetime. However, such tissue growing capacity beyond embryonic development is gradually lost over evolution, suggesting retinal stem cells have become silent in retinas of mammals. But the reason why these RSCs are silenced in homeothemic vertebrates is unknown, which is also an important question in the field of neurogenesis and regeneration. Therefore, a better understanding of RSC niche and the embryonic origin of RSCs of lower vertebrate retinas is critical to understanding why RSCs in higher vertebrates are silent and ultimately may guide activation of dormant RSCs in mammals.
Using zebrafish and cytobow-based clonal analysis, the research team identified the cells in the second or the third layer of the CMZ as the RSCs which were slow-cycling as compared to retina progenitors in CMZ. Moreover, the study also identified a novel set of dormant CMZ tip cells, which may serve as a critical cellular component in the niche.
With precise identification of RSCs, the researchers further investigated the embryonic origin of RSCs using a photoconvertable fluorescent protein Kaede , which could be photoconverted from green to red following exposure to 405 nm laser. Individual cells in the medial epithelial layer at the optic vesicle stage were photoconverted and their clones were traced. According to the lineage analysis, researchers identified a novel population of bipotent embryonic cells in the medial epithelial layer, capable of generating both RSC lineages and RPE lineages. Moreover, these bipotent cells express molecular markers of both RSC and RPE. While the other cells in the ML only could generate retina progenitor cell lineages or pigment cell lineages. All results indicate this newly-identified bipotent cells as the cell origin of retinal stem cells.
This work was carried out by Ms. TANG Xia and Mr. GAO Jianan, under the supervision of Dr. HE Jie, at the Institute of Neuroscience, Chinese Academy of Sciences and CAS Center for Excellence in Brain Science and Intelligence Technology. This work entitled “Bipotent Progenitors as Embryonic Origin of Retinal Stem Cells” was published online in the Journal of Cell Biology on May 2, 2017. It was supported by grant 31471042 from the Natural Science Foundation of China and the Youth Thousand Plan.
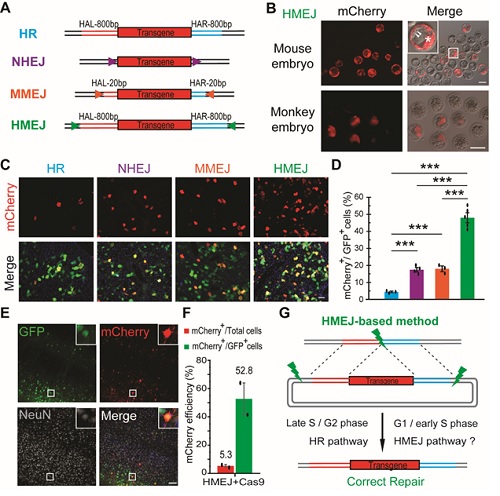
Figure legend: (A) Schematic overview of HR-, NHEJ-, MMEJ- and HMEJ-mediated gene knock-in. HAL/HAR, left/right homology arm; triangles, sgRNA target sites. (B) Representative fluorescence images of HMEJ-mediated gene-edited mouse and monkey embryos at the blastocyst stage. (C) Representative immunofluorescence images of hepatocytes in liver sections at day 7 post injection. Scale bar, 50 μm. GFP, transfected cells. (D) Relative knock-in efficiency measured by the percentage of mCherry+ cells among GFP+ cells. Hepatocytes were harvested at day 7 post injection. The input data points were shown as black dots. ***P < 0.001, unpaired Student’s t-test. (E) Representative immunofluorescence images of neurons in HMEJ-AAV-injected brain sections. Insets, higher magnification images. Scale bar, 100 μm. (F) Relative and absolute knock-in efficiencies measured by the percentage of mCherry+ cells among GFP+ cells or all DAPI+ cells, respectively. At least 2 000 cells of each brain section and three brain sections of each animal were counted. The input data points were shown as black dots. (G) Schematic overview of HMEJ-mediated gene knock-in.

