A new research from the Center for Excellence in Brain Science and Intelligence Technology, CAS, reported on a novel volume imaging and 3D tracking technique that monitors whole brain neural activity in freely swimming larval zebrafish. This study was carried out collaboratively between the Optical Neuroimaging Group led by Dr. WANG Kai at the Institute of Neuroscience (ION) and the Biophysical and Computational Neuroscience group led by Dr. WEN Quan at University of Science and Technology of China (USTC).
In this study, Dr. WANG’s group developed a novel three-dimensional imaging technique called eXtended field-of-view Light Field Microscopy (XLFM) that could image neural activity across the whole brain of larval zebrafish simultaneously and at high speed.
By integrating this imaging technique with a set of high-speed three-dimensional tracking system developed by Dr. WEN’s group, the team successfully carried out rapid whole brain neural activity recording in freely behaving larval zebrafish. For the first time, they captured whole brain neural activity at high spatiotemporal resolution during the prey capture behavior of zebrafish. This work extends traditional experimental paradigms, which can only be performed on head-fixed zebrafish, to freely behaving animals. It opens new window into whole brain dynamics of larval zebrafish in its native and natural environment at unprecedented resolution.
A central goal in systems neuroscience is to understand how distributed neural circuitry dynamics drive animal behaviors. In recent years, larval zebrafish (Danio rerio) have become an increasingly attractive model system to investigate the neural correlates of behaviors owing to their small brain size, optical transparency, and rich behavioral repertoire. Furthermore, applications of advanced optical imaging methods makes it possible to record whole brain neural activity of a movement-restrained larval zebrafish at high spatial-temporal resolution in the virtual reality paradigm. The behavioral repertoire, however, may be further expanded in freely swimming zebrafish whose behavioral states can be directly inferred and when sensory feedback loops are mostly intact and active. Therefore, whole brain imaging in freely swimming zebrafish may allow optical interrogation of brain circuits underlying a range of less explored behaviors.
Whole brain imaging in freely behaving animals has been previously reported in a roundworm Caenorhabditis elegans, by integrating spinning-disk confocal microscopy with a 2D tracking system. However, the significant increase of animal size imposes challenges both in tracking and imaging technologies. To achieve this goal, the team developed two key techniques: (1) a high-speed closed-loop system to retain the entire fish head within the field of view of a high numerical aperture (25×, NA = 1.05) objective in 3D; (2) a new imaging technique XLFM implemented with flashed excitation laser exposure that can image sparse neural activity over the larval zebrafish brain at near single cell resolution and at a volume rate of 77 Hz even when zebrafish moves at a speed up to 10 mm/s. The seamless integration of the tracking and imaging system made it possible to reveal rich whole brain neural dynamics during natural behavior with unprecedented resolution. The team demonstrated the ability of the system during visually evoked and prey capture behaviors in larval zebrafish.
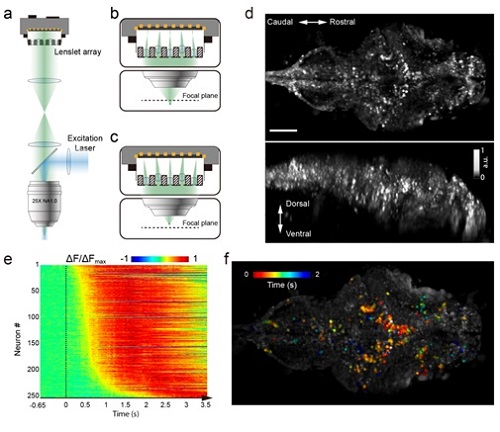
Figure 1. Whole brain imaging of larval zebrafish with XLFM. (a) Schematic of XLFM. Lenslet array position was conjugated to the rear pupil plane of the imaging objective. Excitation laser (blue) provided uniform illumination across the sample. (b–c) Point sources at two different depths formed, through two different groups of micro-lenses, sharp images on the imaging sensor, with positional information reconstructed from these distinct patterns. (d) Maximum intensity projections (MIPs) on time and space of time series volume images of an agarose-restrained larval zebrafish with pan-neuronal nucleus-localized GCaMP6f (huc:h2b-gcamp6f) fluorescence labeling. (e) Normalized neuronal activities of selected neurons exhibited increasing calcium responses after the onset of light stimulation at t = 0. Neurons were ordered by the onset time when the measured fluorescence signals reached 20% of their maximum. (f) Selected neurons in (e) were color coded based on their response onset time. Scale bar is 100 m.
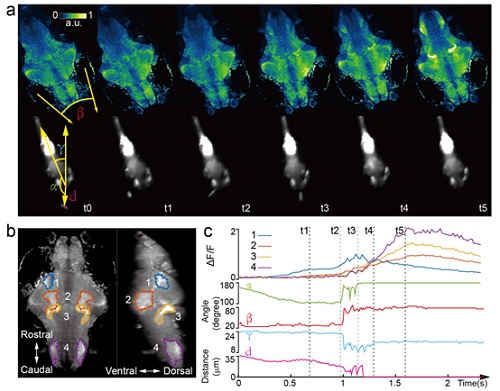
Figure 2. Whole brain imaging of larval zebrafish during prey capture behavior. (a) Renderings of whole brain calcium activity at six time points (up) and the corresponding behavioral images (bottom). Features used to quantify behavior were: fish-paramecium azimuth ?; convergence angle between eyes ?; head orientation ?; and fish-paramecium distance d. (b) Maximum intensity projections of zebrafish brain with pan-neuronal cytoplasm-labeled GCaMP6s (huc:gcamp6s). Boundaries of four brain regions are color marked. (c) Neural dynamics inferred from GCaMP6 fluorescence changes in these four regions during the entire prey capture behavior (up) and the kinematics of behavioral features (bottom). Note that between t2 and t4, fish-paramecium distance d exhibits three abrupt kinks, representing the three attempts to catch prey.
This study entitled "Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio)" has been published online in eLife On September 20, 2017 . This work was conducted collaboratively under the supervision of Dr. WANG Kai, Dr. DU Jiulin from ION and Dr. WEN Quan from USTC. Assistant scientists CONG Lin, HANG Wei from ION and undergraduate WANG Zeguan, graduate student CHAI Yuming from USTC are co-first authors of this paper. This work was supported by the CAS Strategic Priority Research Program of the Chinese Academy of Sciences (XDB02060012), National Science Foundation of China (NSFC-31471051), China Youth Thousand Talents Program (to KW), and CAS Hundreds of Talents Program of China (QW).

