Over 100 different types of RNA modifications have been re-identified genome-wide in transcriptomes. Although accumulated lined of evidence have begun to show their important roles in gene expression regulation, very little is known about the crosstalk between these distinct RNA modifications. Among them, adenosine-to-inosine (A-to-I) and N6-methyladenosines (m6A) are two of the most abundant RNA modifications, and both occur on A bases.
Research teams led by Dr. YANG Li at CAS-MPG Partner Institute for Computational Biology, Chinese Academy of Sciences and Dr. CHEN Ling-Ling at Shanghai Institute of Biochemistry and Cell Biology demonstrate a previously under-appreciated interplay between m6A modification and A-to-I editing, highlighting a complex epitranscriptomic landscape.
The catalytic mechanisms of m6A and A-to-I modifications are distinct. A-to-I conversion is catalyzed by adenosine deaminases acting on RNA (ADARs) that preferentially bind to double-stranded RNA substrates. In contrast, m6A is reversibly processed by a different set of enzymes, and mainly occurs in single-stranded RNA regions. Thus, the different sequence and structure features for A-to-I or m6A suggest that these two chemical modifications do not likely compete for the same A bases. However, an intriguing but unanswered question was whether m6A and A-to-I are always independently regulated.
In this current study, researchers first showed a global A-to-I difference between m6A-positive and m6A-negative RNAs transcribed from the same gene loci. The preferentially presence of A-to-I editing in m6A-negative RNA transcripts suggests a negative correlation of m6A and A-to-I. Knocking down m6A ‘writers’ or ‘eraser’ results in a global alteration of A-to-I editing. Mechanistically, inhibition of m6A modification increases the association of m6A-depleted transcripts with ADAR enzymes for A-to-I editing. This result thus suggests that the unfavorable ADAR1 binding to m6A-transcripts may account for the negative correlation between m6A and A-to-I.
The paper entitled “N6-methyladenosines modulate A-to-I RNA editing” was published in Molecular Cell (Vol 69, Page 126-135, 2018). This study was supported by the grants from Chinese Academy of Sciences, National Natural Science Foundation of China, Ministry of Science and Technology and HHMI International Research Scholar Program.
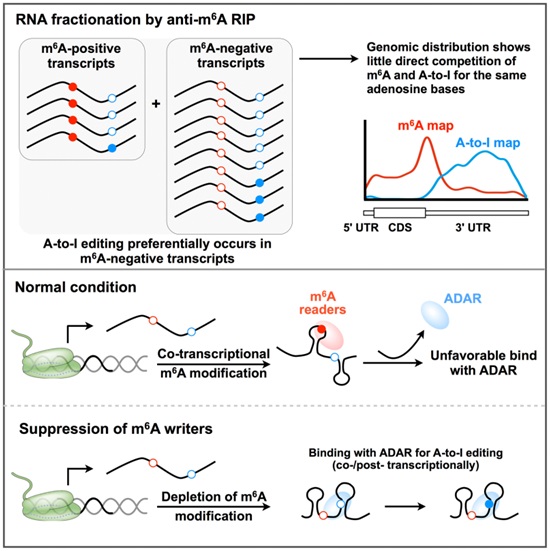
Figure:Negative regulation of m6A modification on A-to-I RNA editing
(Image provided by Dr. YANG Li’s Group)
Contact:
Dr. YANG Li
Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Email: liyang@picb.ac.cn

