Structural variation (SV) in the human genome is defined as a large segment of DNA sequence change, including Deletion, Duplication, Insertion, etc. Many important genetic diseases, including cancer, autism, Alzheimer’s disease, and Parkinson’s disease, were found to be associated with SVs, which is raising a lot of concerns from doctors and geneticists. Due to the great diversity of SVs among different ethnic groups worldwide, as well as the limitation of technology, there is a lack of high-quality data resources concerning representative samples, especially for East Asians.
Prof. XU Shuhua’s group from Fudan University, Prof. ZHANG Guoqing’s group from Shanghai Institute of Nutrition and Health (SINH) of Chinese Academy of Sciences, and Prof. FAN Shaohua’s group from Fudan University jointly published a new SV database, PGG.SV (https://www.biosino.org/pggsv) in Nucleic Acids Research on Oct. 16th, 2022, entitled “PGG.SV: a whole-genome-sequencing-based structural variant resource and data analysis platform”.
This study aims to provide effective guidance and assistance to researchers in related fields by constructing a representative and diverse database of genomic structural variants in global populations. This work is a part of efforts of the Han100K Initiative (https://pog.fudan.edu.cn/han100k).
The research team integrated large-scale sequencing data, including 6,048 whole-genome sequencing data from 177 representative regions and ethnic groups around the world, especially, for the first time, covering 50 ethnic groups in China. In terms of data quality, researchers generated and collected 1,030 long-read sequencing genomes, and applied long-read data to build an SV database for the first time, which offered greater advantages in SV detection.
The database provides user-friendly query functions, including the precise presentation and frequency differences in genomic positions of different ethnic groups, and the relationship between SV and other variants such as Single Nucleotide Variation (SNV). In addition, PGG.SV provides rich clinical effect analysis functions, prediction and enrichment analysis of potential phenotypes, and functions of SV for users with clinical research and other needs. Visualization functions are also provided to search for the SV position on the human genome, and display the structure change in detail for SVs.
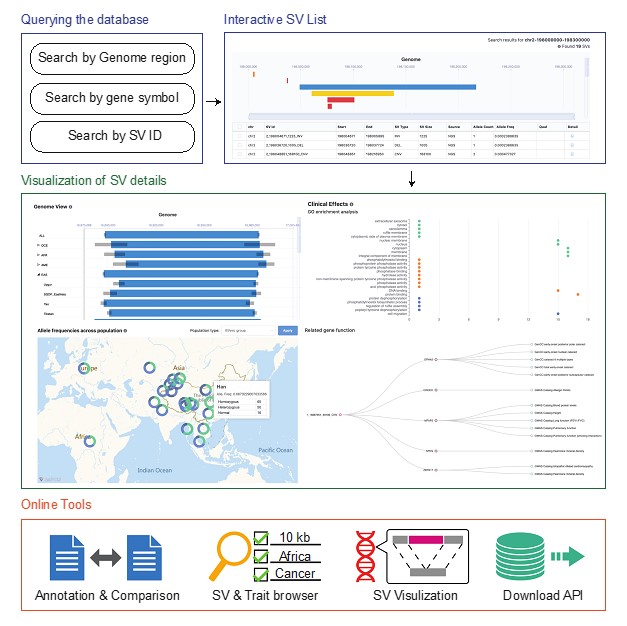
PGG.SV interface. (Image provided by Prof. XU’s group)
Ph.D. student WANG Yimin from SINH, Dr. LING Yunchao from Bio-Med Big Data Center, SINH and Ph.D. student GONG Jiao from Fudan University were joint first authors. Prof. XU Shuhua, Prof. ZHANG Guoqing, and Prof. FAN Shaohua were co-corresponding authors. This study was supported by the Basic Science Center Program, the Strategic Priority Research Program of CAS, the National Natural Science Foundation of China, the UK Royal Society-Newton Advanced Fellowship, Shanghai Municipal Science and Technology Major Project, etc.
Media Contact:
WANG Jin (Ms.)
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: wangjin01@sinh.ac.cn
Web: http://english.sinh.cas.cn/

