Researchers present an innovative network-science inspired method to detect which cell-types change their abundance in aging and disease
Underpinning aging and disease are subtle shifts in cell-type abundance within complex tissues. However, detecting these shifts can be challenging. Single-cell omic data offers the unprecedented opportunity to detect these alterations in cell-type frequency, but doing so requires careful statistical analyses.
Prof. Andrew TESCHENDORFF and Dr. Alok MAITY, a postdoc in his lab from the Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences (CAS), have devised an innovative computational strategy called ELVAR that significantly improves the sensitivity of detecting such shifts in cell-type abundance from single-cell or single-nucleus RNA-Seq data. The key conceptual breakthrough was the realization that more relevant cell-states can be identified using a cell clustering algorithm that takes cell-attributes into account, for instance, their age or the disease stage they derive from. Analogous advances were made in the field of network-science generally, where node-attribute aware clustering has led to improved interpretation of complex datasets from all fields of science. The work from TESCHENDORFF and MAITY shows that similar improvements can be achieved in the realm of single-cell omic data. The authors applied ELVAR to many different single-cell omic datasets, demonstrating in one application, an increased fraction of enterocyte stem-cells and T-regulatory cells in polyps preceding colorectal adenoma, indicating that these shifts in cell-type abundance could be an important contributor and indicator of colorectal cancer risk. In other applications, they found decreased olfactory sensory neurons in the olfactory epithelium of Covid-19 patients experiencing long-term smell loss, and age-related shifts in macrophage polarization within lung-tissue.
What makes this study significant from a biological and clinical perspective, is the ability of ELVAR to detect the cell-type abundance shifts in colon-tissue from sparse single-nucleus RNA-Seq data, paving the way to develop single-cell based early detection and cancer-risk prediction tools from fresh-frozen specimens.
The work was published in Nature Communications under the title “Cell-attribute aware community detection improves differential abundance testing from single-cell RNA-Seq data.” on 5th June 2023. (DOI: 10.1038/s41467-023-39017-z)
The work is funded by the Chinese Academy of Sciences and National Natural Science Foundation of China, grant numbers 31970632 and 32170652.
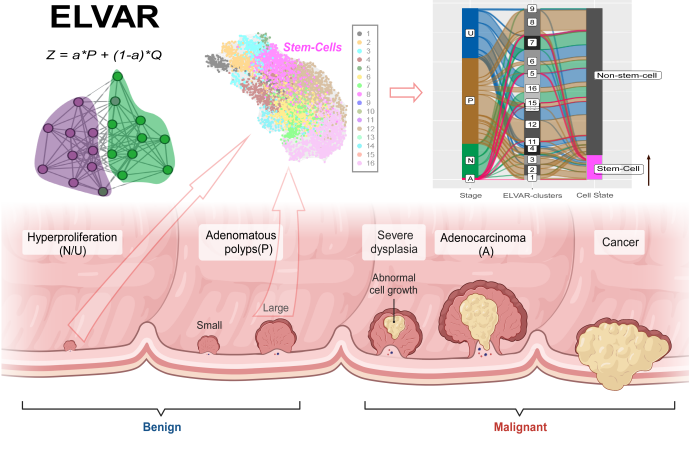
The ELVAR algorithm can be applied to single-nucleus RNA-Seq data from different stages in colorectal adenocarcinoma development, to infer differential abundance of key cell-types, as shown here for enterocyte stem-cells, whose abundance increases in polyps relative to the normal unaffected state.
(Image provided by Prof. TESCHENDORFF 's lab)
Media Contact:
WANG Jin
Shanghai Institute of Nutrition and Health,
Chinese Academy of Sciences
Email: wangjin01@sinh.ac.cn
Web: http://english.sinh.cas.cn/
https://www.nature.com/articles/s41467-023-39017-z

